Research
Active Projects
Biology of bats and their viruses
Bats have a unique ability to tolerate infections with deadly viruses like Ebola, Nipah, SARS, and rabies. But they’re also not a monolith: there are over 1,470 species worldwide, with a wide range of ecosystem roles and unique immune system adaptations. Until recently, comparative immunology with non-model organisms was nearly impossible. However, we’ve developed a unique approach that allows us to characterize bat immune systems — and make predictions based on those data.
To characterize bat immune systems in nature, we’ve collected tens of thousands of samples for proteomic analysis. So far, samples have been collected by team members and collaborators in twelve countries (Australia, Belize, Bulgaria, Costa Rica, Ecuador, Japan, Kenya, Mexico, Oman, Spain, Uganda, and the United States). We’ve also set up cell lines from four bat species so far, and are using in vitro experiments to characterize the immune response to infection. By combining these approaches with genomic data, we’re able to infer how bat immune systems could differ — and train machine learning models to predict which viruses they might be able to host. We test those predictions by working with museums around the world to test samples and characterize novel viruses — and use our algorithms to assess the risk that those viruses could someday pose to human health.
Read more
NSF BII (2022-2027)
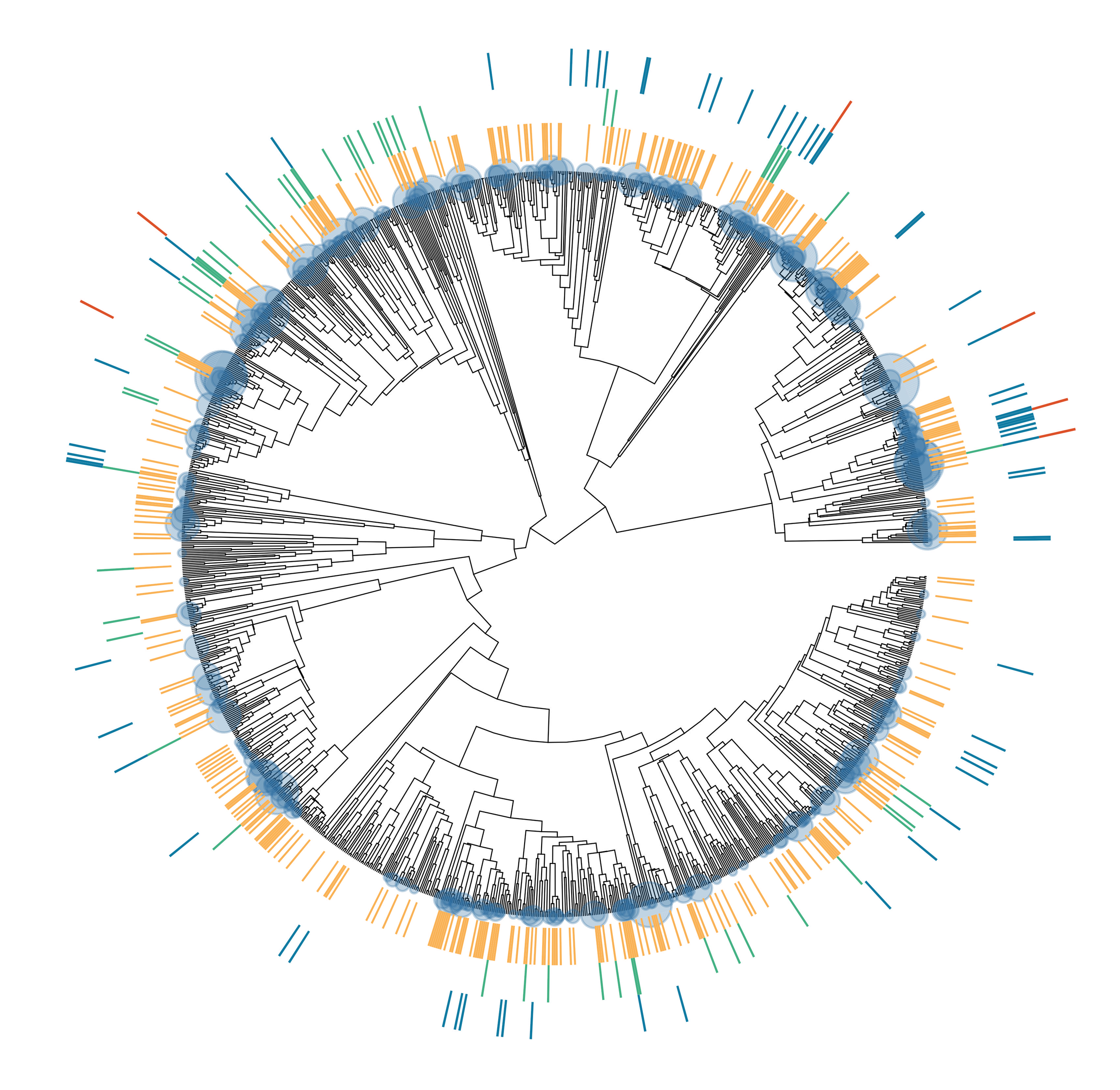
Our project covers the entire bat tree of life, including databases (yellow), field sampling for immune data (green), viral discovery from museum collections (blue), and in vitro experiments with bat cell lines (red). Our efforts are focused on the major reservoirs of zoonotic diseases (circles).
Biology of arboviruses and their vectors
Climate change and deforestation are increasing the risk of future epidemics of pathogens like West Nile, yellow fever, and Zika virus: as temperatures rise, more of the world is at risk of disease transmission, plus viral evolution is accelerating at higher temperatures; mosquitoes and ticks are also on the move.
Our team is using large-scale experimental data and mosquito genomics to understand the evolutionary underpinnings of vector competence, and to develop machine learning models models that can predict virus life cycles and thermal biology in advance of major epidemics. We’re also pioneering an innovative new approach that uses barcoded viruses to study viral evolution within mosquitoes, with a focus on understanding the upper limits of thermal adaptation.
Read more
NSF BII (2022-2027)
The network of mosquito (yellow)-arbovirus (blue) pairings that have been tested by researchers in a laboratory (i.e., vector competence experiments). Bias towards model organisms and medically-important viruses create problems for inference about locally-important vectors or viruses — or future risks.
Climate change and the next pandemic
Our group is leading the effort to understand connections between two of the greatest planetary challenges: climate change and pandemics. Our approach focuses on understanding mechanisms of ecosystem change, and contextualizing potential changes in a world that is already 1.2°C warmer due to human activities.
Our work has shown that climate change will drive dramatic geographic range shifts, especially in bats — producing hotspots of cross-species transmission that coincide with human population centers, especially in south and east Asia. Bats will also be exposed to unprecedented temperatures in their native geographic range, with unpredictable consequences for their health (and for viral spillover).
Ongoing projects include: documenting ongoing range shifts in disease reservoirs; estimating the marginal increase in pandemic risk from both bat coronaviruses and avian influenza; and experiments testing how vertebrate immune systems (and so capacity to tolerate virulent viruses) handle heat-humidity interactions.
Read more
NSF BII (2022-2027)
Climate change is creating new opportunities for pathogens like Ebola virus to jump between wildlife species — creating new disease reservoirs in unexpected places.
Concluded Projects
Mapping hotspots of bat coronavirus diversity
Where will the next Covid-19 get started? Places where people live alongside bat reservoirs of betacoronaviruses (βCoVs) like SARS-CoV-2 — and where surveillance of human outbreaks and animal viruses is limited — are usually at the greatest risk.
Within the first few weeks of the COVID-19 pandemic, our team built an ensemble of machine learning models to identify sampling priorities for new βCoVs. As virologists tested bat samples from around the world for SARS-related viruses, we were able to update our predictions in real-time, and benchmark model performance. Over nearly two years, we found that our best models correctly identified >75% of newly discovered hosts — a key proof of concept for model-guided viral discovery.
Our models predict that over 400 species of bats could be hosts of undiscovered βCoVs, primarily in southeast Asia and equatorial Africa. Many of the top predicted hosts are horseshoe bats (Rhinolophus), the reservoirs of SARS-CoV-1/2 — suggesting that plenty is left to learn about SARS-like viruses. Dozens of species in Europe and the Americas are also predicted to host undiscovered viruses.
To understand where these hotspots came from, we developed a new methodology that maps viral evolutionary potential based on the Geographic Mosaic Theory of Coevolution. Applying this method to bat evolutionary history correctly predicts hotspots of βCoV diversification in the Middle East and Southeast Asia—but also points to new potential hotspots, such as Madagascar and south Asia. Divergent lineages of MERS-like viruses are also expected to be circulating in the Americas.
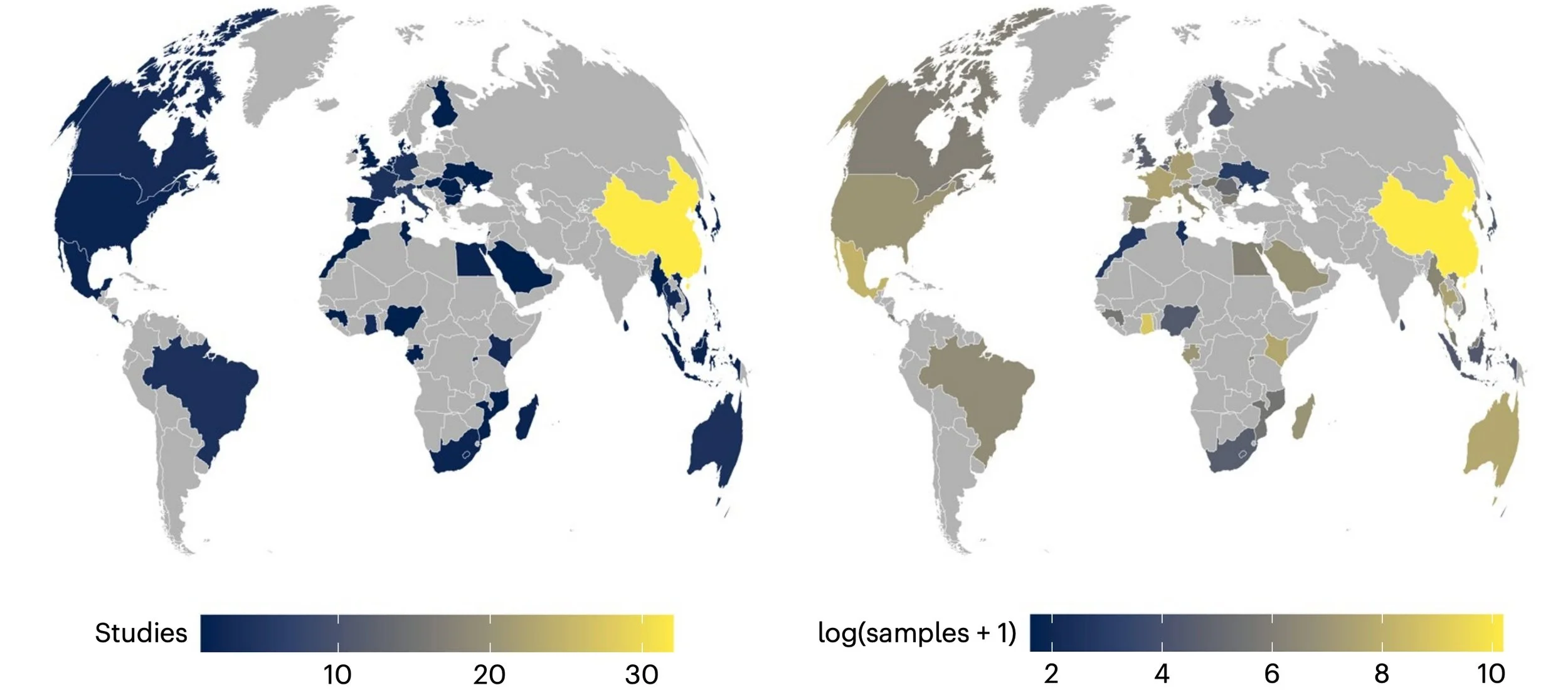
To better direct future surveillance, we developed a comprehensive database of coronavirus sampling between the SARS outbreak and the Covid-19 pandemic. Of the nearly 90,000 samples that have been tested in that time, the vast majority come from China. In the future, the greatest risk of surprise outbreaks will be in under-monitored hotspots of viral diversity like India and central Africa, and even potentially central America and the Andes. Today, we’re working with teams around the world to fill surveillance gaps and test samples based on our priority list.
Read more
IVADO (2020-2024)
Geographic and taxonomic hotspots of undiscovered βCoVs hosts.
Global sampling effort for bat coronaviruses.
Yellow fever dynamics in city-living primates
Where does yellow fever go when it stops circulating in humans? Led by Betânia Drumond (Universidade Federal de Minas Gerais), this project investigated yellow fever dynamics in non-human primates after the 2017 epidemic in Brazil.
Yellow fever was most prevalent in howler monkeys living in intact habitats, but transmission sometimes bled over into urban areas. Viremia in primates decreases in inter-epidemic periods — a tantalizing clue about the molecular mechanisms that allow the virus to persist at low levels for months at a time.
Read more
NIAID CREID Pilot Grant (2021-2023)
Yellow fever dynamics in non-human primates in Brazil (2017-2019).